
Seqinfo object with 25 sequences (1 circular) from an unspecified genome no seqlengths: We can query this information more carefully using an accessor function called We can see thisĪt the end of the output for the GRanges object. This object has an ability of be aware of the chromosomes, or sequences space, on which the features exist. Like many of the objects we will work with for genomic analysis (see GRanges above), Gtf_file <- "/home/cws/CompGenomics/dm6/dm6.Ens_98.gtf" # It could be this easy # Get gene features # We can get them from UCSC # txdb <- makeTxDbFromUCSC(genome="mm10", tablename="ensGene") # We've done our analysis thus far with a particular version of ensembl # in the form of a GTF file, so let's use that.
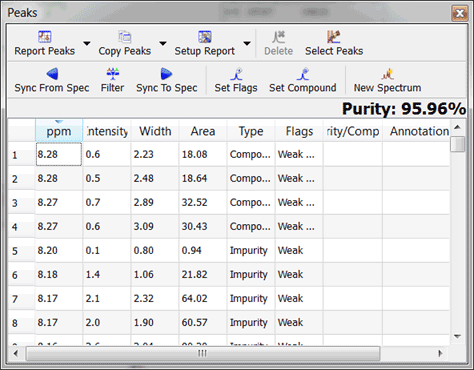
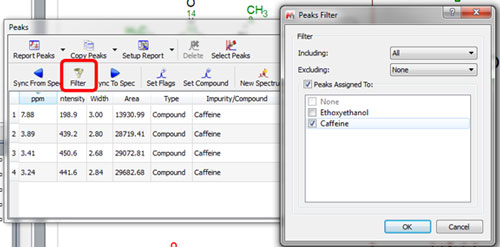
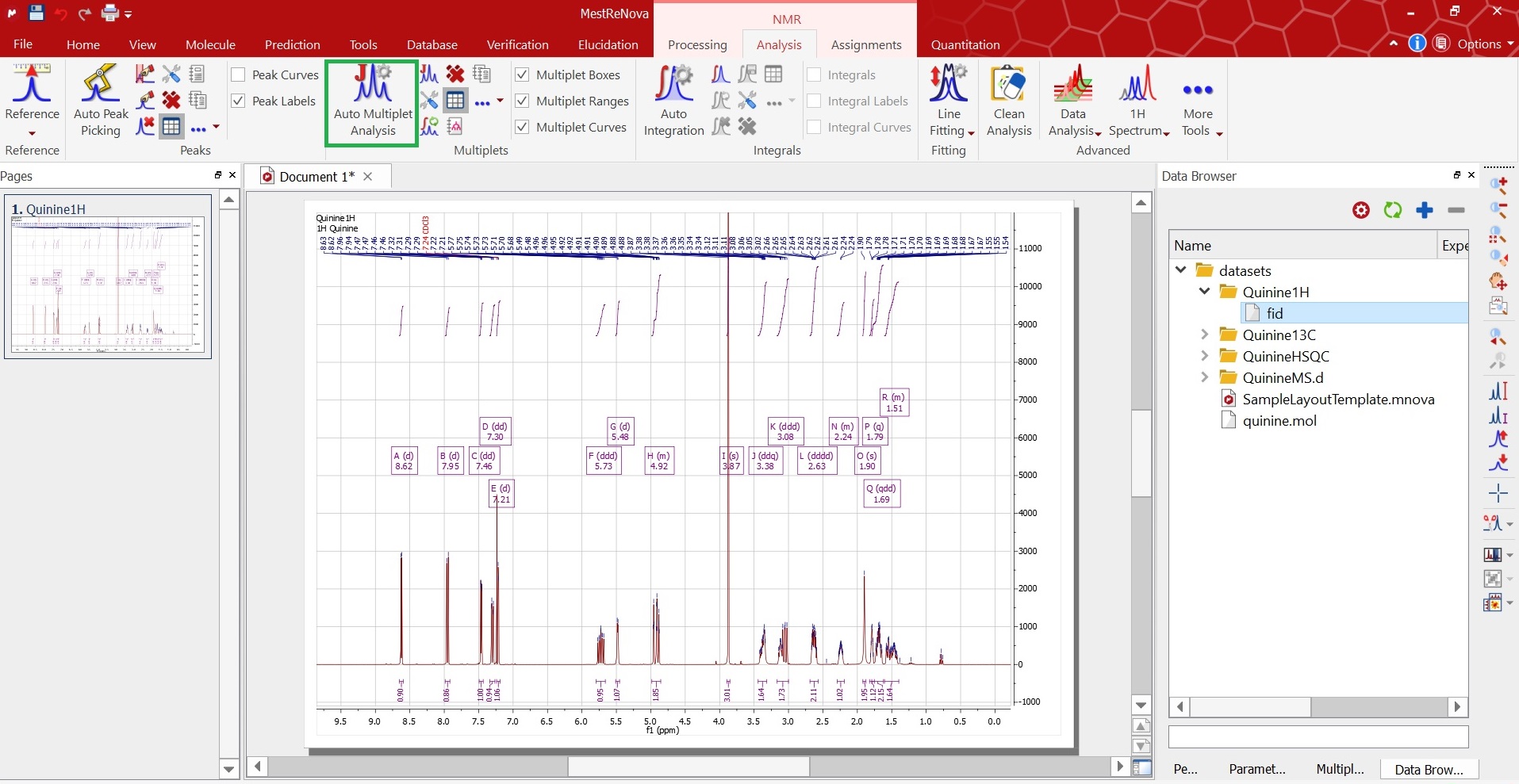
HOW TO ASSIGN PEAKS IN MESTRENOVA CODE
1H NMR spectrum of ethanol with normalized integral numbers.# CODE BLOCK #2 # Load the library library(GenomicFeatures) Two peaks in a ratio of 1H:2H could correspond to one and two hydrogens, or they could correspond to two and four hydrogens, etc.įigure NMR19. These numbers could correspond to numbers of hydrogens, or simply to their lowest common factors. They are reduced to a lowest common factor so that their ratios are easier to compare. Source: Spectrum taken in CDCl 3 on a Varian Gemini 2000 Spectrometer with 300 MHz Oxford magnet. 1H NMR spectrum of ethanol with numerical integrals. Often, instead of displaying raw data, the integrals are measured and their heights are displayed on the spectrum.įigure NMR18. 1H NMR spectrum of ethanol with broken integral line. Sometimes, the integral line is cut into separate integrals for each peak so that they can be compared to each other more easily.įigure NMR17. The area is related to the amount of radio waves absorbed at that frequency, and the amount of radio waves absorbed is proportional to the number of hydrogen atoms absorbing the radio waves.
This measurement is shown as a jump or step upward in the integral line the vertical distance that the line rises is proportional to the area of the peak. Where the line crosses the frequency of a peak, the area of the peak is measured. In raw form, an integral is a horizontal line running across the spectrum from left to right. Integral data can be given in different forms. integration reveals the ratio of one type of hydrogen to another within a molecule.That information helps narrow down the number of possible structures of the sample, and so it makes structure elucidation of an unknown sample much easier. You can also see by integration that there are three hydrogens of one type, two of the second type, and one of the third type - corresponding to the CH 3 or methyl group, the CH 2 or methylene group and the OH or hydroxyl group. Looking at the spectrum of ethanol, you can see that there are three different kinds of hydrogens in the molecule. 1H NMR spectrum of ethanol with solid integral line.
That in turn shows that the ratio of the hydrogen atoms in the three different environments is 1:2:3.įigure NMR16. You measure the height gained at each peak or group of peaks by measuring the distances shown in green in the diagram above - and then find their ratio.įor example, if the heights were 0.7 cm, 1.4 cm and 2.1 cm, the ratio of the peak areas would be 1:2:3. The height gained is proportional to the area under the peak or group of peaks. When the integrator trace crosses a peak or group of peaks, it gains height. In the diagram, the integrator trace is shown in red.Īn integrator trace measures the relative areas under the various peaks in the spectrum. An integrator trace (or integration trace) can be used to find the ratio of the numbers of hydrogen atoms in different environments in an organic compound.Īn integrator trace is a computer generated line which is superimposed on a proton NMR spectra. Chemical shift can show how many different types of hydrogens are found in a molecule integration reveals the number of hydrogens of each type.

There is additional information obtained from 1H NMR spectroscopy that is not typically available from 13C NMR spectroscopy.


 0 kommentar(er)
0 kommentar(er)
